Tutorial: Easy Machine Learning with Latent Variable Models in AMIDST¶
In AMIDST toolbox 0.4.2 the module latent-variable-models, that contains a wide range of predefined latent variable models (see table below), was included. In this tutorial we will show how the use of this module simplifies the learning and inference processes. In fact, you will be able to learn your model and to perform inference with a few lines of code.
Besides of the simplicity, the required code for learning a latent variable model is also flexible: you will be able to change the learnt model or the inference algorithm just with some slight modfications in the code. Another advantage of using AMIDST for learning one of the predefined models is that the procedure is traparent to the format of your training data: you will use the same methods regardless of learning from a local or a distributed dataset (with Flink). Note that this last feature was included in the version 0.5.0 of the toolbox.
Setting up¶
In order to follow this tutorial, you will need to have the java 8 (i.e. SDK 1.8 or higher) installed in your system. For more details about the system requirements, see this link. Additionally, you can download a ready-to-use IntelliJ maven project with all the code examples in this tutorial. For that, use the following command:
$ git clone https://github.com/amidst/tutorial.git
Alternativelly, you would rather create a new maven project or use an existing one. For that, you might check the Getting Started page.
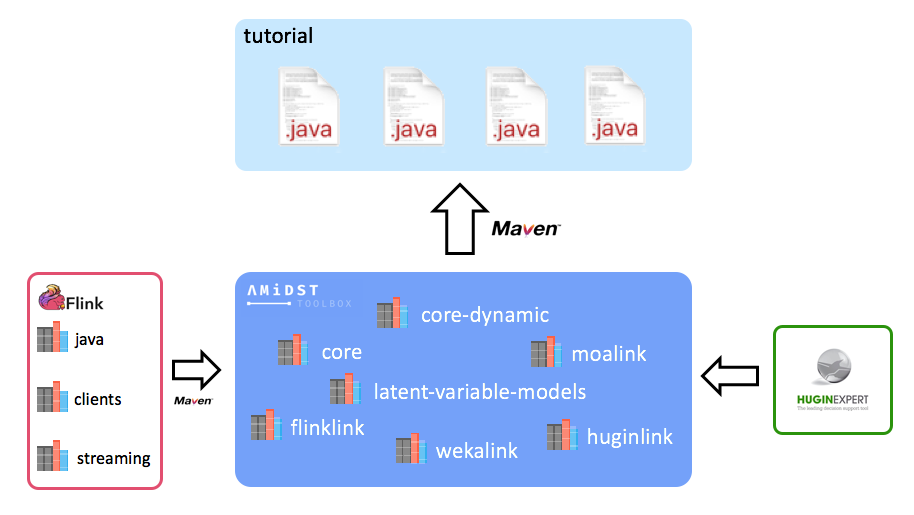
Note that this project does not contain any AMIDST source or binary, it only has some .java files using the AMIDST functionallity. Instead, each of the AMIDST modules are provided through maven. Doing that, the transitive dependences of the AMIDST toolbox are also downloaded in a transparent way for the user. An scheme of this idea is shown below:
If we open the downloaded project, we will see that it contains the following relevant folders and files:
- datasets: folder with local and distributed datasets used in this tutorial in ARFF format.
- doc: folder containing documentation about this tutorial.
- lib: folder for storing those libraries not available through maven.
- src/main/java: folder with all the code example.
- pom.xml: this is the maven configuration file where the AMIDST dependencies are defined.
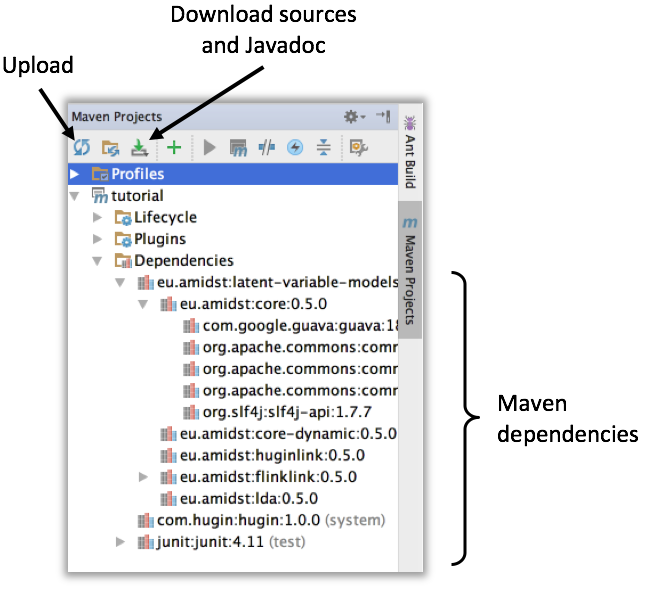
In the pom.xml file of the downloaded project, only the module called latent-variable-models is linked. However some other AMIDST are also loaded as latent-variable-models module depends on them. This is the case of the modules called core, core-dynamic, flinklink, etc. You can see the full list of dependencies in the maven project panel, usually located on the right side of the window (see image below). If dependencies are not automatically downloaded, click on Upload button. It is recommended to download the sources and java
Static Models¶
Learning and saving to disk¶
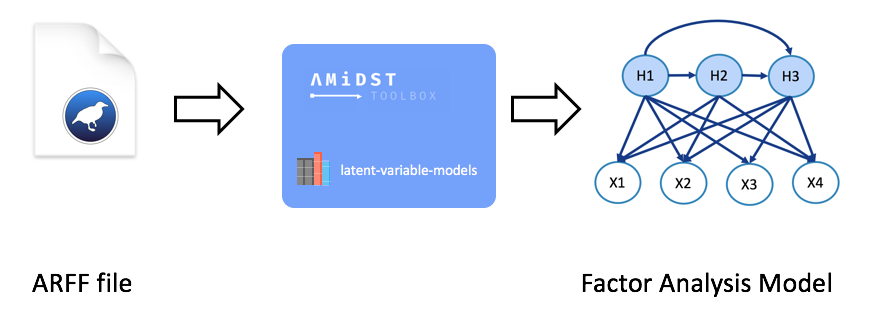
Here we will see how can we learnt a static model from a local dataset (non-distributed). In particular, we will use the financial-like dataset datasets/simulated/cajamar.arff containing 4 continuous (normal distributed) variables. From this data, a Factor Analysis model will be learnt. In short, this model aims to reduce the dimensionality of a set of observed continuous variables by expressing them as combination of gaussians. A synthesis of this process is shown in the image below where: \(X1, X2, X3\) and \(X4\) are the observed variables and \(H1, H2\) and \(H3\) are the latent variables representing the combination of gaussians.
Note that the required functionality for learning the predefined model is provided by the module latent-variable-models. A code-example for learning a factor analysis model is shown below.
package eu.amidst.tutorial.usingAmidst.examples;
import COM.hugin.HAPI.ExceptionHugin;
import eu.amidst.core.datastream.DataInstance;
import eu.amidst.core.datastream.DataStream;
import eu.amidst.core.io.BayesianNetworkWriter;
import eu.amidst.core.io.DataStreamLoader;
import eu.amidst.core.models.BayesianNetwork;
import eu.amidst.huginlink.io.BayesianNetworkWriterToHugin;
import eu.amidst.latentvariablemodels.staticmodels.FactorAnalysis;
import eu.amidst.latentvariablemodels.staticmodels.MixtureOfFactorAnalysers;
import eu.amidst.latentvariablemodels.staticmodels.Model;
import java.io.IOException;
/**
* Created by rcabanas on 23/05/16.
*/
public class StaticModelLearning {
public static void main(String[] args) throws ExceptionHugin, IOException {
//Load the datastream
String filename = "datasets/simulated/cajamar.arff";
DataStream<DataInstance> data = DataStreamLoader.open(filename);
//Learn the model
Model model = new FactorAnalysis(data.getAttributes());
// ((MixtureOfFactorAnalysers)model).setNumberOfLatentVariables(3);
model.updateModel(data);
BayesianNetwork bn = model.getModel();
System.out.println(bn);
// Save with .bn format
BayesianNetworkWriter.save(bn, "networks/simulated/exampleBN.bn");
// Save with hugin format
//BayesianNetworkWriterToHugin.save(bn, "networks/simulated/exampleBN.net");
}
}
For learning any of the available static models, we create an object of any of the classes inheriting from the class Model. These classes encapsulates all the fuctionality for learning/updating a latent-variable model. For example, in the code above we create an object of the class FactorAnalysis which is actually stored as Model object. The flexibility of the toolbox is due to this hierarchical desing: if we aim to change the model learnt from our data, we simply have to change the constructor used (assuming that our data also fits the constraints of the new model). For example, if we aim to learn a mixture of factor analysers instead, we simply have to replace the line
Model model = new FactorAnalysis(data.getAtributes());
by
Model model = new MixtureOfFactorAnalysers(data.getAtributes());
Note that the method for learning the model, namely Model::updateMode(DataStream<DataInstance>) will always be the same regardless of the particular type of static model.
The actual learnt model is an object of the class BayesianNetwork which is stored as a member variable of Model. Thus, for using the network, we simply have to extract with the method Model::getModel(). One of the actions we can perform with it is saving it into the local file system. For saving it in .bn format:
BayesianNetworkWriter::save(BayesianNetwork bn, String path)
Alternatively, and assuming that we have the hugin library available, we can also save it in .net format:
BayesianNetworkWriteToHuginr::save(BayesianNetwork bn, String path)
Learning from Flink¶
In previous section we showed how the AMIDST toolbox can be used for learning a static model from a non-distributed dataset. In addition, you can use the pre-defined models to process massive data sets in a distributed computer cluster using Apache Flink. In particular, the model can be learnt from a distributed ARFF folder or from a file accesible via a HDFS url. A scheme of the learning process is shown below.
A code example for learning from Flink is shown below. Note that it only differs from the one in previous section in lines 25 to 33. In these lines, the Flink session is configurated and the stream is loaded, which is managed with an object of the class DataFlink (instead of DataStream).
package eu.amidst.tutorial.usingAmidst.examples;
import COM.hugin.HAPI.ExceptionHugin;
import eu.amidst.core.datastream.DataInstance;
import eu.amidst.core.io.BayesianNetworkWriter;
import eu.amidst.core.models.BayesianNetwork;
import eu.amidst.flinklink.core.data.DataFlink;
import eu.amidst.flinklink.core.io.DataFlinkLoader;
import eu.amidst.latentvariablemodels.staticmodels.FactorAnalysis;
import eu.amidst.latentvariablemodels.staticmodels.Model;
import eu.amidst.tutorial.usingAmidst.Main;
import org.apache.flink.api.java.ExecutionEnvironment;
import org.apache.flink.configuration.Configuration;
import java.io.IOException;
/**
* Created by rcabanas on 23/05/16.
*/
public class StaticModelFlink {
public static void main(String[] args) throws IOException, ExceptionHugin {
//Set-up Flink session.
// Configuration conf = new Configuration();
// conf.setInteger("taskmanager.network.numberOfBuffers", 12000);
final ExecutionEnvironment env = ExecutionEnvironment.getExecutionEnvironment();
//env.getConfig().disableSysoutLogging();
// env.setParallelism(Main.PARALLELISM);
//Load the datastream
String filename = "datasets/simulated/cajamarDistributed.arff";
DataFlink<DataInstance> data = DataFlinkLoader.open(env, filename, false);
//Learn the model
Model model = new FactorAnalysis(data.getAttributes());
model.updateModel(data);
BayesianNetwork bn = model.getModel();
System.out.println(bn);
// Save with .bn format
BayesianNetworkWriter.save(bn, "networks/simulated/exampleBN.bn");
// Save with hugin format
//BayesianNetworkWriterToHugin.save(bn, "networks/simulated/exampleBN.net");
}
}
In previous example, the distributed dataset is stored in our local file system. Instead, we might need to load from a distributed file system. For that, simply replace the string indicating the path. That is, replace
String filename = "datasets/simulated/cajamarDistributed.arff"
by
String filename = "hdfs://distributed-server/path-to-file"
Inference¶
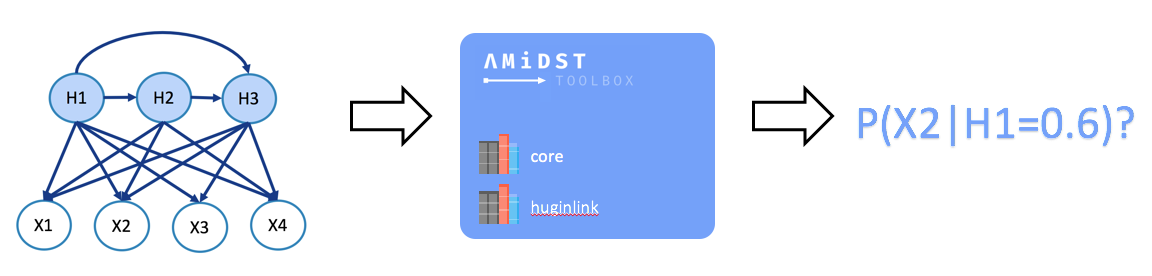
Making probabilistic inference in BNs (a.k.a belief updating) consists of the computation of the posterior probability distribution for a set of variables of interest given some evidence over some other variables (see image below).
The inference process is the same regardless of the way we have learnt our model: we simply have to obtain the BN learnt (stored as an object of the class BayesianNetwork), set the target variables and the evidence. As an example, let us consider the following code:
package eu.amidst.tutorial.usingAmidst.examples;
import eu.amidst.core.distribution.Distribution;
import eu.amidst.core.inference.InferenceAlgorithm;
import eu.amidst.core.inference.messagepassing.VMP;
import eu.amidst.core.io.BayesianNetworkLoader;
import eu.amidst.core.models.BayesianNetwork;
import eu.amidst.core.variables.Assignment;
import eu.amidst.core.variables.HashMapAssignment;
import eu.amidst.core.variables.Variable;
import eu.amidst.core.variables.Variables;
import java.io.IOException;
/**
* Created by rcabanas on 23/05/16.
*/
public class StaticModelInference {
public static void main(String[] args) throws IOException, ClassNotFoundException {
BayesianNetwork bn = BayesianNetworkLoader.loadFromFile("networks/simulated/exampleBN.bn");
Variables variables = bn.getVariables();
//Variabeles of interest
Variable varTarget = variables.getVariableByName("LatentVar1");
Variable varObserved = null;
//we set the evidence
Assignment assignment = new HashMapAssignment(2);
varObserved = variables.getVariableByName("Income");
assignment.setValue(varObserved,0.0);
//we set the algorithm
InferenceAlgorithm infer = new VMP(); //new HuginInference(); new ImportanceSampling();
infer.setModel(bn);
infer.setEvidence(assignment);
//query
infer.runInference();
Distribution p = infer.getPosterior(varTarget);
System.out.println("P(LatentVar1|Income=0.0) = "+p);
//Or some more refined queries
System.out.println("P(0.7<LatentVar1<6.59 |Income=0.0) = " + infer.getExpectedValue(varTarget, v -> (0.7 < v && v < 6.59) ? 1.0 : 0.0 ));
}
}
Note that the learning algorithm can be easily changed by simply modifying line 35 where VMP algorithm. If we aim to use Importance Sampling algorithm, replace such line with:
InferenceAlgorithm infer = new ImportanceSampling();
Alternatively, we can use Hugin Inference algorithm (assuming that we have the corresponding libraries):
InferenceAlgorithm infer = new HuginInference();
Custom static model¶
It could happend that your model of interest is not predifined. In that case you can implement it yourself. For that purpose, create a new class inheriting the class Model. Then, add the code to the constructor with an object Attributes as input parameter, and the code of the method void buildDAG(). This last method is called before learning process and creates the object representing the DAG. As an example, the code below shows how to create a custom Gaussian Mixture.
package eu.amidst.tutorial.usingAmidst.practice;
import eu.amidst.core.datastream.Attributes;
import eu.amidst.core.datastream.DataInstance;
import eu.amidst.core.datastream.DataStream;
import eu.amidst.core.io.DataStreamLoader;
import eu.amidst.core.models.BayesianNetwork;
import eu.amidst.core.models.DAG;
import eu.amidst.core.variables.Variable;
import eu.amidst.core.variables.Variables;
import eu.amidst.latentvariablemodels.staticmodels.FactorAnalysis;
import eu.amidst.latentvariablemodels.staticmodels.Model;
import eu.amidst.latentvariablemodels.staticmodels.exceptions.WrongConfigurationException;
/**
* Created by rcabanas on 23/05/16.
*/
public class CustomGaussianMixture extends Model{
Attributes attributes;
public CustomGaussianMixture(Attributes attributes) throws WrongConfigurationException {
super(attributes);
this.attributes=attributes;
}
@Override
protected void buildDAG() {
/** Create a set of variables from the given attributes**/
Variables variables = new Variables(attributes);
/** Create a hidden variable with two hidden states*/
Variable hiddenVar = variables.newMultinomialVariable("HiddenVar",2);
//We create a standard naive Bayes
DAG dag = new DAG(variables);
for (Variable variable: variables){
if (variable==hiddenVar)
continue;
dag.getParentSet(variable).addParent(hiddenVar);
}
//This is needed to maintain coherence in the Model class.
this.dag=dag;
this.vars = variables;
}
//Method for testing the custom model
public static void main(String[] args) {
String filename = "datasets/simulated/cajamar.arff";
DataStream<DataInstance> data = DataStreamLoader.open(filename);
//Learn the model
Model model = new CustomGaussianMixture(data.getAttributes());
model.updateModel(data);
BayesianNetwork bn = model.getModel();
System.out.println(bn);
}
}
Dynamic Models¶
Learning and saving to disk
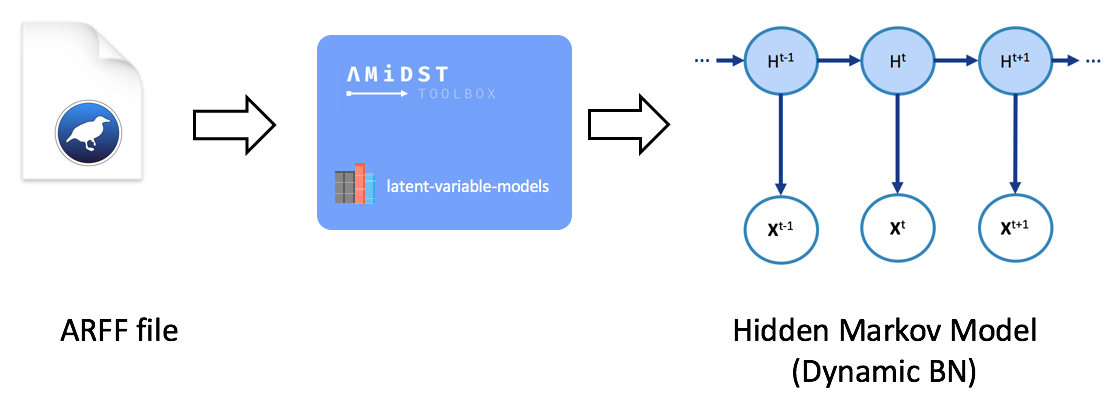
When dealing with temporal data, it might be advisable to learn a dynamic model. The module latent-variable-models in AMDIST also suports such kinds of models. For that the classes inheriting DynamicModel will be used. A synthesis of the learning process is shown below.
A code-example is given below. Here, a Hidden Markov Model (HMM) is learnt from a financial dataset with temporal information. Note the difference within the static learning is the way the dataset is loaded: now, it is handled with an object of the class DataStream<DynamicDataInstance>. Finally, the DBN learnt is saved to disk with the method DynamicBayesianNetworkWriter::save(String pathFile).
package eu.amidst.tutorial.usingAmidst.examples;
import COM.hugin.HAPI.ExceptionHugin;
import eu.amidst.core.datastream.DataStream;
import eu.amidst.dynamic.datastream.DynamicDataInstance;
import eu.amidst.dynamic.io.DynamicBayesianNetworkWriter;
import eu.amidst.dynamic.io.DynamicDataStreamLoader;
import eu.amidst.dynamic.models.DynamicBayesianNetwork;
import eu.amidst.latentvariablemodels.dynamicmodels.DynamicModel;
import eu.amidst.latentvariablemodels.dynamicmodels.HiddenMarkovModel;
import java.io.IOException;
/**
* Created by rcabanas on 23/05/16.
*/
public class DynamicModelLearning {
public static void main(String[] args) throws IOException, ExceptionHugin {
//Load the datastream
String filename = "datasets/simulated/cajamar.arff";
DataStream<DynamicDataInstance> data = DynamicDataStreamLoader.loadFromFile(filename);
//Learn the model
DynamicModel model = new HiddenMarkovModel(data.getAttributes());
model.updateModel(data);
DynamicBayesianNetwork dbn = model.getModel();
System.out.println(dbn);
// Save with .bn format
DynamicBayesianNetworkWriter.save(dbn, "networks/simulated/exampleDBN.dbn");
// Save with hugin format
//DynamicBayesianNetworkWriterToHugin.save(dbn, "networks/simulated/exampleDBN.net");
}
}
Inference¶
In the following code-example, the inference process of dynamic models is illustrated. First, a DBN is loaded from disk (line 24). Then, a dynamic dataset is loaded for testing our model (lines 29 to 30). Then the inference algorithm and target variables are set. In the final loop, the inference is perform for each data instance.
package eu.amidst.tutorial.usingAmidst.examples;
import eu.amidst.core.datastream.DataStream;
import eu.amidst.core.distribution.UnivariateDistribution;
import eu.amidst.core.inference.ImportanceSampling;
import eu.amidst.core.variables.Variable;
import eu.amidst.dynamic.datastream.DynamicDataInstance;
import eu.amidst.dynamic.inference.FactoredFrontierForDBN;
import eu.amidst.dynamic.inference.InferenceAlgorithmForDBN;
import eu.amidst.dynamic.io.DynamicBayesianNetworkLoader;
import eu.amidst.dynamic.io.DynamicDataStreamLoader;
import eu.amidst.dynamic.models.DynamicBayesianNetwork;
import java.io.IOException;
/**
* Created by rcabanas on 23/05/16.
*/
public class DynamicModelInference {
public static void main(String[] args) throws IOException, ClassNotFoundException {
DynamicBayesianNetwork dbn = DynamicBayesianNetworkLoader.loadFromFile("networks/simulated/exampleDBN.dbn");
System.out.println(dbn);
//Testing dataset
String filenamePredict = "datasets/simulated/cajamar.arff";
DataStream<DynamicDataInstance> dataPredict = DynamicDataStreamLoader.open(filenamePredict);
//Select the inference algorithm
InferenceAlgorithmForDBN infer = new FactoredFrontierForDBN(new ImportanceSampling()); // new ImportanceSampling(), new VMP(),
infer.setModel(dbn);
Variable varTarget = dbn.getDynamicVariables().getVariableByName("discreteHiddenVar");
UnivariateDistribution posterior = null;
//Classify each instance
int t = 0;
for (DynamicDataInstance instance : dataPredict) {
if (instance.getSequenceID()>0)
break;
infer.addDynamicEvidence(instance);
infer.runInference();
posterior = infer.getFilteredPosterior(varTarget);
System.out.println("t="+t+", P(discreteHiddenVar | Evidence) = " + posterior);
posterior = infer.getPredictivePosterior(varTarget, 2);
//Display the output
System.out.println("t="+t+"+5, P(discreteHiddenVar | Evidence) = " + posterior);
t++;
}
}
}
Note that the inference algorithm can be easily change if line 33 is modified by replacing it with:
InferenceAlgorithmForDBN = new FactoredFrontierForDBN(new VMP());
Custom dynamic model¶
Like for static models, we might be interested in creating our own dynamic models. In this case, you will have to create a class inheriting DynamicModel. Here below an example of a custom Kalman Filter is given.
package eu.amidst.tutorial.usingAmidst.practice;
import COM.hugin.HAPI.ExceptionHugin;
import eu.amidst.core.datastream.Attributes;
import eu.amidst.core.datastream.DataInstance;
import eu.amidst.core.datastream.DataStream;
import eu.amidst.core.io.DataStreamLoader;
import eu.amidst.core.models.BayesianNetwork;
import eu.amidst.core.variables.Variable;
import eu.amidst.dynamic.datastream.DynamicDataInstance;
import eu.amidst.dynamic.io.DynamicBayesianNetworkWriter;
import eu.amidst.dynamic.io.DynamicDataStreamLoader;
import eu.amidst.dynamic.models.DynamicBayesianNetwork;
import eu.amidst.dynamic.models.DynamicDAG;
import eu.amidst.dynamic.variables.DynamicVariables;
import eu.amidst.latentvariablemodels.dynamicmodels.DynamicModel;
import eu.amidst.latentvariablemodels.dynamicmodels.HiddenMarkovModel;
import eu.amidst.latentvariablemodels.dynamicmodels.KalmanFilter;
import eu.amidst.latentvariablemodels.staticmodels.FactorAnalysis;
import eu.amidst.latentvariablemodels.staticmodels.Model;
import eu.amidst.latentvariablemodels.staticmodels.exceptions.WrongConfigurationException;
import java.io.IOException;
import java.util.ArrayList;
import java.util.List;
/**
* Created by rcabanas on 23/05/16.
*/
public class CustomKalmanFilter extends DynamicModel {
Attributes attributes;
public CustomKalmanFilter(Attributes attributes) throws WrongConfigurationException {
super(attributes);
this.attributes=attributes;
}
@Override
protected void buildDAG() {
/*number of continuous hidden variable*/
int numHiddenVars=3;
/** Create a set of dynamic variables from the given attributes**/
variables = new DynamicVariables(attributes);
/* List of continuous hidden vars*/
List<Variable> gaussianHiddenVars = new ArrayList<>();
for(int i=0; i<numHiddenVars; i++) {
Variable Hi = variables.newGaussianDynamicVariable("gaussianHiddenVar" + i);
gaussianHiddenVars.add(Hi);
}
DynamicDAG dynamicDAG = new DynamicDAG(this.variables);
for (Variable h : gaussianHiddenVars) {
dynamicDAG.getParentSetTimeT(h).addParent(h.getInterfaceVariable());
}
for (Variable variable: variables) {
if (gaussianHiddenVars.contains(variable))
continue;
for (Variable h : gaussianHiddenVars) {
dynamicDAG.getParentSetTimeT(variable).addParent(h);
}
}
//This is needed to maintain coherence in the DynamicModel class.
this.variables = variables;
this.dynamicDAG = dynamicDAG;
}
public static void main(String[] args) throws IOException, ExceptionHugin {
//Load the datastream
String filename = "datasets/simulated/cajamar.arff";
DataStream<DynamicDataInstance> data = DynamicDataStreamLoader.loadFromFile(filename);
//Learn the model
DynamicModel model = new CustomKalmanFilter(data.getAttributes());
model.updateModel(data);
DynamicBayesianNetwork dbn = model.getModel();
System.out.println(dbn);
}
}